s01: Pandas
Contents
s01: Pandas#

This is a quick introduction to Pandas.
Objectives of this session:#
Learn simple and some more advanced usage of pandas dataframes
Get a feeling for when pandas is useful and know where to find more information
Understand enough of pandas to be able to read its documentation.
Pandas is a Python package that provides high-performance and easy to use data structures and data analysis tools. This page provides a brief overview of pandas, but the open source community developing the pandas package has also created excellent documentation and training material, including:
a Getting started guide - including tutorials and a 10 minute flash intro
thorough Documentation
a cookbook.
Let’s get a flavor of what we can do with pandas. We will be working with an example dataset containing the passenger list from the penguins, which is often used in Kaggle competitions and data science tutorials. First step is to load pandas:
import pandas as pd # pd is the standard abbreviation
import numpy as np
We can download the data from seaborn directly reading into a DataFrame:
import seaborn as sns
pengs = sns.load_dataset("penguins")
We can now view the dataframe to get an idea of what it contains and print some summary statistics of its numerical data:
# print the first 5 lines of the dataframe
pengs.head()
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | Male |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | Female |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | Female |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | Female |
What’s in a dataframe?#
Clearly, pandas dataframes allows us to do advanced analysis with very few commands, but it takes a while to get used to how dataframes work so let’s get back to basics.
As we saw above, pandas dataframes are a powerful tool for working with tabular data. A pandas pandas.DataFrame is composed of rows and columns:
Lets get some detailed information about the numerical data of the df.
# print summary statistics for each column
pengs.describe()
| bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | |
|---|---|---|---|---|
| count | 342.000000 | 342.000000 | 342.000000 | 342.000000 |
| mean | 43.921930 | 17.151170 | 200.915205 | 4201.754386 |
| std | 5.459584 | 1.974793 | 14.061714 | 801.954536 |
| min | 32.100000 | 13.100000 | 172.000000 | 2700.000000 |
| 25% | 39.225000 | 15.600000 | 190.000000 | 3550.000000 |
| 50% | 44.450000 | 17.300000 | 197.000000 | 4050.000000 |
| 75% | 48.500000 | 18.700000 | 213.000000 | 4750.000000 |
| max | 59.600000 | 21.500000 | 231.000000 | 6300.000000 |
Ok, so we have information on pengouins data. With the summary statistics we see that the body mass is 4201 g, maximum flipper length is 231 mm, etc.
Let’s say we’re interested in the mean boddy mass per species. With two one-liners, we can find the average body mass and plot corresponding histograms (pandas.DataFrame.groupby(), pandas.DataFrame.hist()):
print(pengs.groupby("species")["body_mass_g"].mean())
species
Adelie 3700.662252
Chinstrap 3733.088235
Gentoo 5076.016260
Name: body_mass_g, dtype: float64
pengs.hist(column='body_mass_g', bins=25)
array([[<AxesSubplot:title={'center':'body_mass_g'}>]], dtype=object)
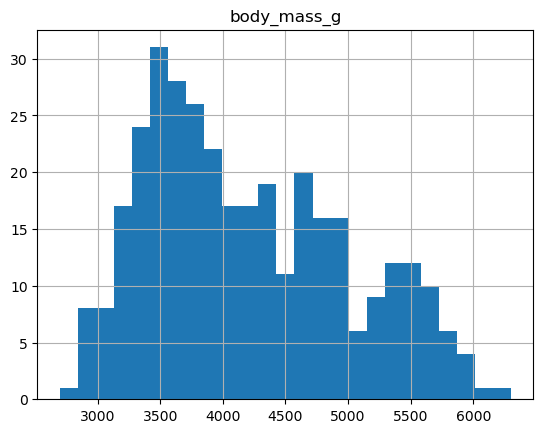
groupby is a powerful method which splits a dataframe and aggregates data in groups. We start by creating a new column child to indicate whether a pengouin was a child or not, based on the existing body_mass_g column. For this example, let’s assume that you are a child when you weight less than 4500 g:
pengs["child"] = pengs["body_mass_g"] < 4000
Now we can test the if the flipper length is different for childs by grouping the data on species and then creating further sub-groups based on child:
pengs.groupby(["species", "child"])["flipper_length_mm"].mean()
species child
Adelie False 195.282051
True 188.098214
Chinstrap False 203.062500
True 193.596154
Gentoo False 217.262295
True 208.000000
Name: flipper_length_mm, dtype: float64
Each column of a dataframe is a pandas.Series object - a dataframe is thus a collection of series:
# print some information about the columns
pengs.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 344 entries, 0 to 343
Data columns (total 8 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 species 344 non-null object
1 island 344 non-null object
2 bill_length_mm 342 non-null float64
3 bill_depth_mm 342 non-null float64
4 flipper_length_mm 342 non-null float64
5 body_mass_g 342 non-null float64
6 sex 333 non-null object
7 child 344 non-null bool
dtypes: bool(1), float64(4), object(3)
memory usage: 19.3+ KB
Now we can already see what columns are present in the df. Any easier way jsut to display the column names is to use the function df.columns.
pengs.columns
Index(['species', 'island', 'bill_length_mm', 'bill_depth_mm',
'flipper_length_mm', 'body_mass_g', 'sex', 'child'],
dtype='object')
Unlike a NumPy array, a dataframe can combine multiple data types, such as numbers and text, but the data in each column is of the same type. So we say a column is of type int64 or of type object.
Let’s inspect one column of the penguin data (first downloading and reading the datafile into a dataframe if needed, see above):
pengs["flipper_length_mm"]
pengs.flipper_length_mm # same as above
0 181.0
1 186.0
2 195.0
3 NaN
4 193.0
...
339 NaN
340 215.0
341 222.0
342 212.0
343 213.0
Name: flipper_length_mm, Length: 344, dtype: float64
Now let us adress not just a single column, but also include the rows as well. Adressing a row with numbers is what Pandas calls the index:
pengs.index
RangeIndex(start=0, stop=344, step=1)
We saw above how to select a single column, but there are many ways of selecting (and setting) single or multiple rows, columns and values. We can refer to columns and rows either by number or by their name (loc, iloc, at, iat):
pengs.loc[0,"island"] # select single value by row and column
pengs.loc[:10,"bill_length_mm":"body_mass_g"] # slice the dataframe by row and column *names*
pengs.iloc[0:2,3:6] # same slice as above by row and column *numbers*
pengs.at[0,"flipper_length_mm"] = 42 # set single value by row and column *name* (fast)
pengs.at[0,"species"] # select single value by row and column *name* (fast)
pengs.iat[0,5] # select same value by row and column *number* (fast)
pengs["is_animal"] = True # set a whole column
Dataframes also support boolean indexing, just like we saw for numpy arrays:
idx_big = pengs["body_mass_g"] > 4500
pengs[idx_big]
| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | child | is_animal | |
|---|---|---|---|---|---|---|---|---|---|
| 7 | Adelie | Torgersen | 39.2 | 19.6 | 195.0 | 4675.0 | Male | False | True |
| 39 | Adelie | Dream | 39.8 | 19.1 | 184.0 | 4650.0 | Male | False | True |
| 45 | Adelie | Dream | 39.6 | 18.8 | 190.0 | 4600.0 | Male | False | True |
| 81 | Adelie | Torgersen | 42.9 | 17.6 | 196.0 | 4700.0 | Male | False | True |
| 101 | Adelie | Biscoe | 41.0 | 20.0 | 203.0 | 4725.0 | Male | False | True |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 338 | Gentoo | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | Female | False | True |
| 340 | Gentoo | Biscoe | 46.8 | 14.3 | 215.0 | 4850.0 | Female | False | True |
| 341 | Gentoo | Biscoe | 50.4 | 15.7 | 222.0 | 5750.0 | Male | False | True |
| 342 | Gentoo | Biscoe | 45.2 | 14.8 | 212.0 | 5200.0 | Female | False | True |
| 343 | Gentoo | Biscoe | 49.9 | 16.1 | 213.0 | 5400.0 | Male | False | True |
115 rows × 9 columns
Using the boolean index idx_big, we can select specific row from an entire df based on values from a single column.
Tidy data#
The above analysis was rather straightforward thanks to the fact that the dataset is tidy.
In short, columns should be variables and rows should be measurements, and adding measurements (rows) should then not require any changes to code that reads the data.
What would untidy data look like? Here’s an example from some run time statistics from a 1500 m running event:
runners = pd.DataFrame([
{'Runner': 'Runner 1', 400: 64, 800: 128, 1200: 192, 1500: 240},
{'Runner': 'Runner 2', 400: 80, 800: 160, 1200: 240, 1500: 300},
{'Runner': 'Runner 3', 400: 96, 800: 192, 1200: 288, 1500: 360},
])
runners.head()
| Runner | 400 | 800 | 1200 | 1500 | |
|---|---|---|---|---|---|
| 0 | Runner 1 | 64 | 128 | 192 | 240 |
| 1 | Runner 2 | 80 | 160 | 240 | 300 |
| 2 | Runner 3 | 96 | 192 | 288 | 360 |
runners = pd.melt(runners, id_vars="Runner",
value_vars=[400, 800, 1200, 1500],
var_name="distance",
value_name="time"
)
runners.head()
| Runner | distance | time | |
|---|---|---|---|
| 0 | Runner 1 | 400 | 64 |
| 1 | Runner 2 | 400 | 80 |
| 2 | Runner 3 | 400 | 96 |
| 3 | Runner 1 | 800 | 128 |
| 4 | Runner 2 | 800 | 160 |
In this form it’s easier to filter, group, join and aggregate the data, and it’s also easier to model relationships between variables.
The opposite of melting is to pivot data, which can be useful to view data in different ways as we’ll see below.
For a detailed exposition of data tidying, have a look at this article.
Creating dataframes from scratch#
We saw above how one can read in data into a dataframe using the read_csv() function. Pandas also understands multiple other formats, for example using read_excel, read_json, etc. (and corresponding methods to write to file: to_csv, to_excel, to_json, etc.)
But sometimes you would want to create a dataframe from scratch. Also this can be done in multiple ways, for example starting with a numpy array (see DataFrame docs
Let’s create a df for our study and fill it with random numbers which do not make any sense at all.
import random
import string
n = 6
ids = [''.join(random.choices(string.ascii_uppercase + string.digits, k=5)) for i in range(0,n)] # this line generates IDs from Uppercase letters A-Z and Number 0-9 of length 5
nms_cols = ['age','height','MoCA']
rand_age = np.random.randint(18,99,n)
rand_height = np.random.randint(150,200,n)
rand_moca = np.random.randint(0,30,n)
df = pd.DataFrame(list(zip(rand_age, rand_height, rand_moca)), index=ids, columns=nms_cols)
df.head()
| age | height | MoCA | |
|---|---|---|---|
| W6YCR | 96 | 170 | 29 |
| V7W9X | 50 | 189 | 25 |
| 0IY04 | 41 | 190 | 18 |
| 2JTTH | 22 | 189 | 15 |
| O71H4 | 84 | 199 | 15 |

